Your brain and spinal cord are suspended in about 125 mL of cerebrospinal fluid (CSF) that not only protects your precious neurons like a kind of liquid bubble wrap, but contains a rich mix of proteins that record the activity of the nervous system it guards.
A study led by Washington University researchers has now revealed a unique atlas of proteins linked to the neurodegenerative condition Alzheimer's disease which could also potentially be targeted by medicines.
Alzheimer's is difficult to study because you can't take a good look at someone's brain until after they've died. Most studies that have aimed to identify the genes linked to Alzheimer's risk and protection rely on dead brain tissue, which can only give a limited view of the disease, usually in its latest stages.
Another approach scientists have taken in the past is to study blood plasma. Though blood can provide a convenient measure of some markers of brain disease, it doesn't directly interact with the brain tissues affected by Alzheimer's like CSF does.
Spinal fluid starts out as blood plasma, so it has a lot in common with it, but it contains far fewer proteins and different levels of electrolytes. The proteins that CSF does contain can tell us a lot about the cellular activities within the brain, however, as well as the genes that are responsible for them.
Tracing numerous proteins back to networks of genes that may influence the progress of a disease like Alzheimer's is a challenge that can involve mapping each step of expression from gene to protein.
Genomicist Carlos Cruchaga from Washington University and his collaborators mined two existing datasets based at Washington University for the genetic data and CSF samples of 3,506 people, some with Alzheimer's disease and some without.
This allowed the researchers to determine which cellular pathways – and the genes and proteins they involve – might be throwing spanners into the works for people with Alzheimer's.
"Sometimes within a region of DNA known to be associated with Alzheimer's there are many genes, and we don't know which of those genes are driving the medical condition," Cruchaga says.
"By adding the proteins to the analysis, we can determine the gene driving the association, determine the molecular pathway that they are part of, as well as to identify novel protein-to-protein interactions that otherwise will not be possible."
They also matched proteins in these CFS samples with areas of the human genome that have already been associated with Alzheimer's disease, and determined which of these genes and proteins have the strongest statistical links with the biological pathways that lead to Alzheimer's.
This narrowed the dataset of 6,361 CSF proteins down to just 38 that are likely involved in causing Alzheimer's. Fifteen of these can be targeted by available drugs, some of which have already been associated with a decreased Alzheimer's disease risk. This study shows us why.
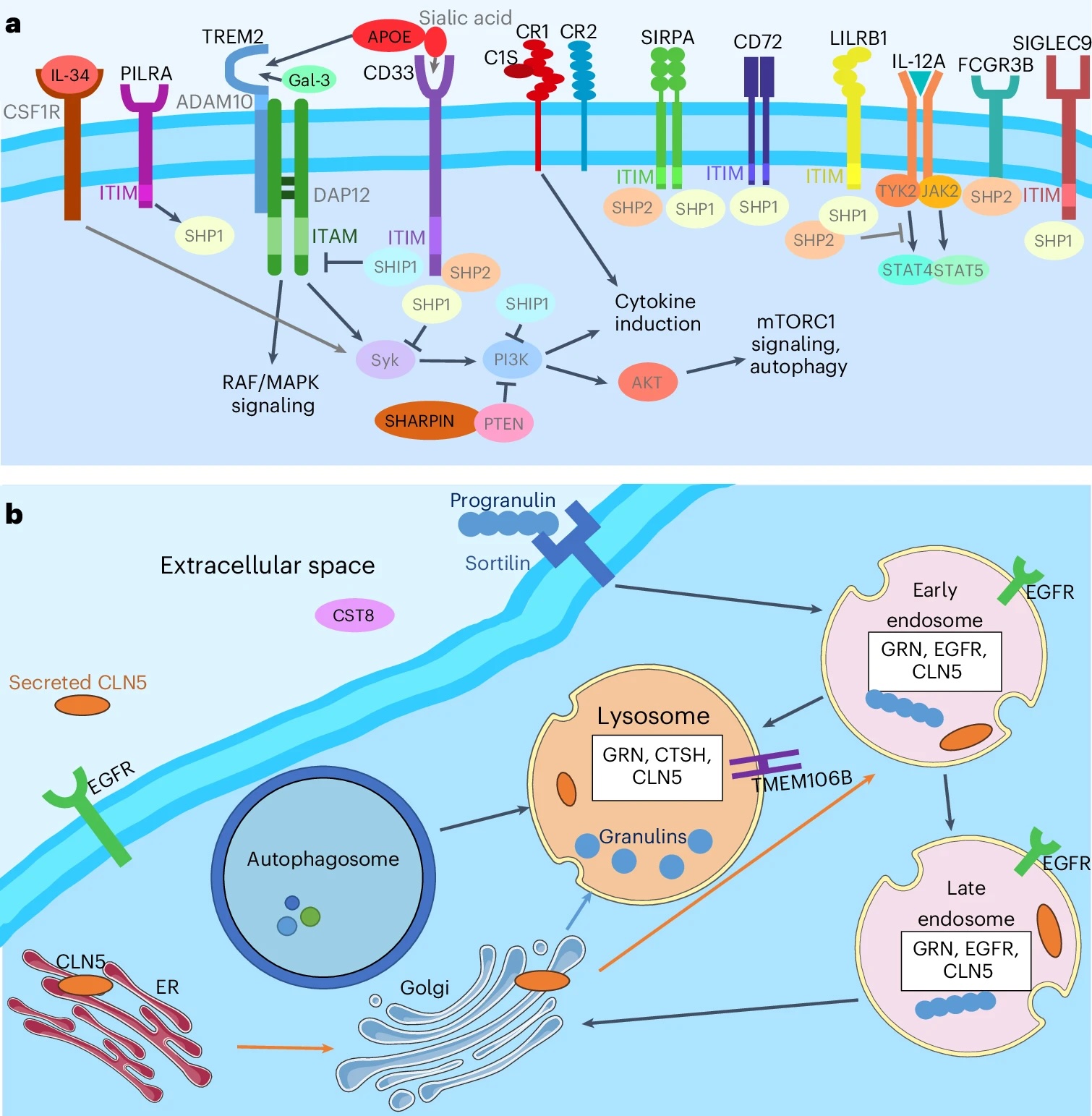
"The novelty and the strength of this analysis is that we have defined proteins that modify risk," Cruchaga says. "So now that we have the causal steps, we can establish where the steps are leading to in the brain."
With all of this information, they were able to develop a proteomics-based model that can predict Alzheimer's disease with more accuracy than existing genetics-based models.
The researchers hope that this application of CSF proteomics could be translated to many neurological conditions, like Parkinson's disease and schizophrenia.
"That's the power of this approach – once you have an atlas of genetic variants, and that of the protein levels, you can apply this to any disease," Cruchaga says.
The study was published in Nature Genetics.