Scientists in the US have developed a powerful new genome editing system that could offer significantly more precision and efficiency than the current CRISPR standard.
While CRISPR-Cas9 is a revolutionary medical technology that lay the modern foundations for editing genetic code – including variants associated with disease – there have long been concerns over its potential for imprecision.
Specifically, many fear that CRISPR-Cas9 editing carries the potential to introduce errors in the form of uncontrolled insertions and deletions in genetic code, called indels.
Researchers say the new system, dubbed 'prime editing' by its inventors at the Broad Institute of MIT and Harvard University, could change the game thanks to a new protein that enables high-precision edits of genetic targets.
"A major aspiration in the molecular life sciences is the ability to precisely make any change to the genome in any location," says genome biologist David Liu from the the Broad Institute.
"We're not aware of another editing technology in mammalian cells that offers this level of versatility and precision with so few byproducts."
The basis of the new prime editing method is an enzyme called reverse transcriptase. The CRISPR system also uses an enzyme – Cas9 – to cut DNA strands, so that alternative genetic code can be inserted.
A 2017 breakthrough pioneered by Liu's lab greatly enhanced the precision of the system, enabling single-letter changes in DNA base pairs instead of replacing entire stretches of code at once.
Now, thanks to reverse transcriptase, which is used in conjunction with Cas9 in prime editing, genomic editing has been upgraded again.
In prime editing, a guide RNA called pegRNA guides a modified form of the Cas9 enzyme to snip only a single strand of DNA (preventing the double-strand breaks that can induce unintended disruptions).
After this, the reverse transcriptase enzyme directly copies edited genetic information contained in the pegRNA to the targeted genomic site. You can see a detailed explanation of the procedure in the infographic below:
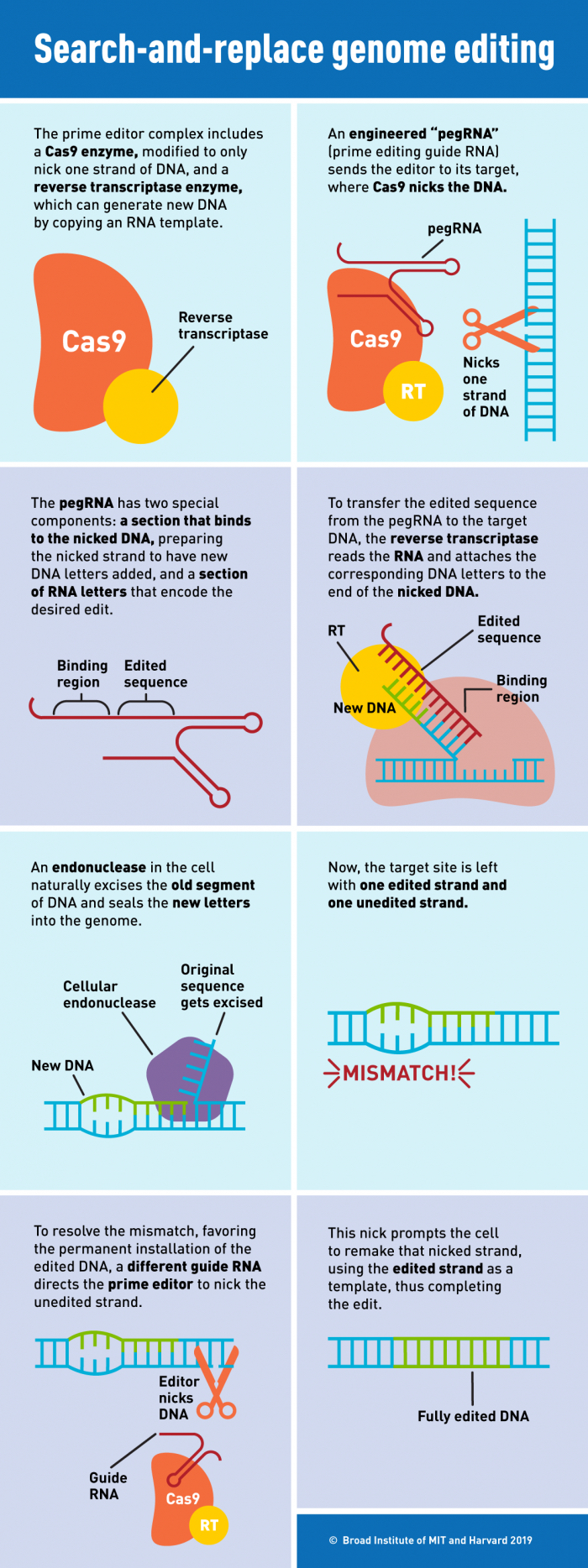 (Broad Institute)
(Broad Institute)
"The versatility of prime editing quickly became apparent as we developed this technology," explains chemical biologist Andrew Anzalone.
"The fact that we could directly copy new genetic information into a target site was a revelation. We were really excited."
The flexibility of the system means that for the first time, researchers can effectively swap one DNA 'letter' with another – among the chemicals adenine (A), cytosine (C), guanine (G), and thymine (T) – in all 12 possible ways.
That's a marked improvement on what the base editor advancements made in 2017 were capable of, meaning entirely new kinds of genetic edits in human diseases are now possible compared to what could be done before.
"With prime editing, we can now directly correct the sickle-cell anaemia mutation back to the normal sequence and remove the four extra DNA bases that cause Tay Sachs disease, without cutting DNA entirely or needing DNA templates," Liu explains.
In the researchers' new paper, the team details those procedures in lab tests, among an account of over 175 edits in human and mouse cells – with results that produced fewer unwanted byproducts, and introduced lower amounts of off-target edits than the Cas9 approach by itself would have created.
It's landmark stuff, but especially given the rapid pace at which these technologies are evolving, the researchers are eager to emphasise we're still only at the beginning of testing the system, to find out what it's capable of in lab environment.
"Much additional research is needed to further understand and improve prime editing in a broad range of cell types and organisms, to assess off-target prime editing in a genome-wide manner, and to further characterise the extent to which prime editors might affect cells," the authors write in their paper.
Nonetheless, it's clear there could be a huge amount of opportunity here in terms of broadening the theoretical and practical scale of genome editing – extending the technique, in principle, to 89 percent of known pathogenic human genetic variants, the researchers say.
The commercial opportunities are also potentially massive. While the researchers will be granting other researchers a non-commercial licence to explore the approach, commercial applications for the patent-pending system, could be more than considerable for its inventors.
For those in the research community, though, there's palpable excitement for some pretty incredible science, and the promise of prime editing.
"It's early days, but the initial results look fantastic," genetics researcher Brittany Adamson from Princeton University, who wasn't involved with the study, told Nature.
"You're going to see a lot of people using it."
The findings are reported in Nature.
