From the spiral arms of galaxies to microscopic snow crystals, nature seems to fall into fractal-like patterns that repeat in increasingly smaller increments. No matter how small you go, parts of the pattern still resemble the whole.
One exception appears to be molecules, which have not been known to exhibit self-similarity at changing scales. That is, until now.
Researchers from Germany, Sweden, and the UK have discovered an enzyme produced by a single-celled organism that can arrange itself into a fractal – not just any fractal, but a repeating pattern of triangles known as a Sierpiński triangle.
The enzyme is a form of citrate synthase produced by the cyanobacterium Synechococcus elongatus, and its evolution from non-fractal precursors suggests that such molecular patterns can emerge surprisingly quickly.
"We stumbled on this structure completely by accident and almost couldn't believe what we saw when we first took images of it using an electron microscope," says biochemist Franziska Sendker of the Max Planck Institute for Terrestrial Microbiology in Germany.
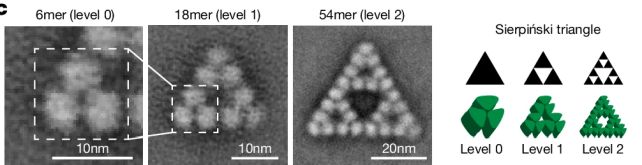
"The protein makes these beautiful triangles and as the fractal grows, we see these larger and larger triangular voids in the middle of them, which is totally unlike any protein assembly we've ever seen before."

There's a reason for that, the researchers found. They used electron microscopy to study the molecule's structure at an atomic level, examining how the protein chains are linked.
Most molecules have a highly symmetrical structure, with each protein chain falling into the same arrangement and relationship to the protein chains around it. This particular structure of citrate synthase manages to break that rule: the protein chains link up in slightly different ways, depending on their position in the molecule.
The result is the Sierpiński arrangement, and, interestingly, the team's research found that it seems to have been a complete accident that has no function whatsoever. When the team genetically manipulated S. elongatus to produce non-fractal citrate synthase, it made no difference to the bacterium at all.
"Self-assembly is often used by evolution to regulate enzymes, but in this case the cyanobacterium that this enzyme is found in does not seem to care much whether or not its citrate synthase can assemble into a fractal," says evolutionary biologist Georg Hochberg of the Max Planck Institute for Terrestrial Microbiology.
"This prompted us to wonder whether this might just be a harmless accident of evolution. Such accidents can happen when the structure in question isn't too difficult to construct."
And it seems that a fractal pattern isn't a very difficult thing for a molecule to achieve. The researchers explored the evolutionary history by using ancestral sequence reconstruction to determine the precursors of the citrate synthase molecule as it appears today, and the amino acid substitutions that could have altered its structure over time.
It turns out that it takes only a very small number of mutations to alter the molecule's shape, with startling rapidity. But easy come is easy go – it can disappear again just as quickly. In fact, the team's research suggests that fractal molecules have emerged and disappeared in different species of cyanobacteria multiple times in the past.
It's just that, if a fractal structure is not biologically helpful, there is no reason for the organism to retain it, so off it goes. For S. elongatus, it's possible that the Sierpiński molecule is only temporary. Or perhaps in nature there is some benefit that was not observed in a laboratory setting.
"We suspect that evolutionary transitions in self-assembly may be more common than structural databases make it seem," the researchers write in their paper.
"Perhaps only a small fraction ever become important to their organisms and persist. Many others will fade as quickly as they appeared. We can therefore only wonder about how many unique assemblies have evolved over the eons that never made it to the present for us to observe."
The findings have been published in Nature.
