In June 2000, two rival groups of researchers shook hands in the shared success of a milestone in biology – the delivery of a rough draft of the human genome.
What started with an incomplete map of our chromosomes has since bloomed into a vast trove of individualized sequences from all corners of the globe, and in many cases stretching far back in time.
Somewhere in that ocean of decoded DNA is a story of our shared humanity.
Unfortunately, reading it is easier said than done. Not only is the sheer mass of data a problem, subtle differences in samples, diverse formats, and analysis techniques prioritizing different kinds of errors all present obstacles to a unified interpretation.
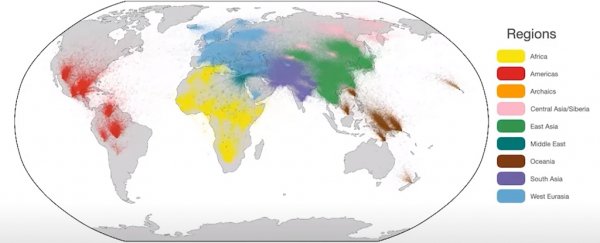
Now researchers from the Big Data Institute (BDI) at the University of Oxford in the UK have made a significant start, by merging a forest of more than 3,600 individual sequences from 215 populations into a single, enormous tree.
The tree's branches comprise of a mind-blowing 231 million ancestral lineages. At its base is a spread of roots represented by eight ancient, highly detailed human genome sequences, with thousands of smaller snippets used to confirm their place deep in our past.
Among them are three Neanderthal genomes, one genome from a Denisovan, and a small family who lived in Siberia more than four thousand years ago.
"Essentially, we are reconstructing the genomes of our ancestors and using them to form a series of linked evolutionary trees that we call a 'tree sequence'," says geneticist Anthony Wilder Wohns, who led the study while completing his doctorate at the BDI.
"We can then estimate when and where these ancestors lived."
Their tree sequence method makes use of what's known as a succinct data structure – a computing concept that aims to represent data in an optimal amount of space that also limits the amount of time needed to probe it all with questions.
We might apply similar thinking when saving files on our own computer, finding a compromise between compressing documents and squeezing them into long lists of folders, or simply saving everything on the desktop.
In this specific case, a tree sequence finds correlations between different branches of a tree to help make the large pools of information easier to study.
By turning the data into graphs with nodes representing various lineages and mapping mutations along the edges, massive genetic databases can not only be squeezed into a relatively small space, but can be accessed more easily by algorithms designed to search for interesting statistics.
"The power of our approach is that it makes very few assumptions about the underlying data and can also include both modern and ancient DNA samples," says Wohns, who further explains their work in the video below.
Incorporating labels on the geographical locations of sequences allowed the team to estimate where certain common ancestors might have once lived and how they moved about.
Not only does this reveal events we already suspect, such as how human populations migrated from Africa, it hints at changes in population densities within ancestral groups we're still learning about, such as the Denisovans.
Thanks to the efficiency of this process, the already impressive tree has plenty of room to grow as more genetic data become available in the future.
Adding millions more genomes will only make any further results more accurate, pinpointing exactly where a novel sequence fits in a genealogy that stretches around the world.
"This genealogy allows us to see how every person's genetic sequence relates to every other, along all the points of the genome," says BDI evolutionary geneticist, Yan Wong.
Thinking even bigger, there's no reason the same approach couldn't be applied to other species, possibly one day contributing to a global tapestry of life on Earth.
"While humans are the focus of this study, the method is valid for most living things; from orangutans to bacteria," says Wohns.
"It could be particularly beneficial in medical genetics, in separating out true associations between genetic regions and diseases from spurious connections arising from our shared ancestral history."
This research was published in Science.