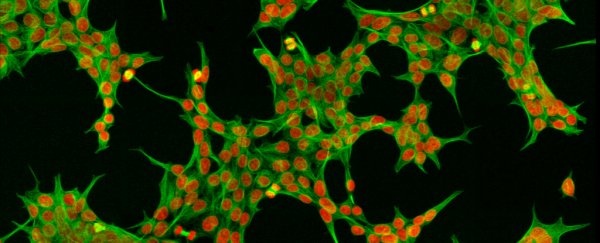
Inside every cell of the human body is a constellation of proteins, millions of them. They're all jostling about, being speedily assembled, folded, packaged, shipped, cut and recycled in a hive of activity that works at a feverish pace to keep us alive and ticking.
But without a full inventory of the protein universe inside our cells, scientists are hard-pressed to appreciate on a molecular level what goes wrong with our bodies that leads to disease.
Now, researchers have developed a new technique that uses artificial intelligence to assimilate data from microscopy images of single cells and biochemical analyses, to create a 'unified map' of subcellular components – half of which, it turns out, we've never seen before.
"Scientists have long realized there's more that we don't know than we know, but now we finally have a way to look deeper," says computer scientist and network biologist Trey Ideker of the University of California (UC) San Diego.
Microscopes, powerful as they are, allow scientists to peer inside single cells, down to the level of organelles such as mitochondria, the power packs of cells, and ribosomes, the protein factories. We can even add fluorescent dyes to easily tag and track proteins.
Biochemistry techniques can go deeper still, honing in on single proteins by using, for example, targeted antibodies that bind the protein, pull it out of the cell, and see what else is attached to it.
Integrating those two approaches is a challenge for cell biologists.
"How do you bridge that gap from nanometer to micron-scale? That has long been a big hurdle in the biological sciences," explains Ideker.
"Turns out you can do it with artificial intelligence – looking at data from multiple sources and asking the system to assemble it into a model of a cell."
The result: Ideker and colleagues have flipped textbook maps of globular cells which give us a birds-eye view of candy-colored organelles into an intricate web of protein-protein interactions, organized by the teensy distances between them.
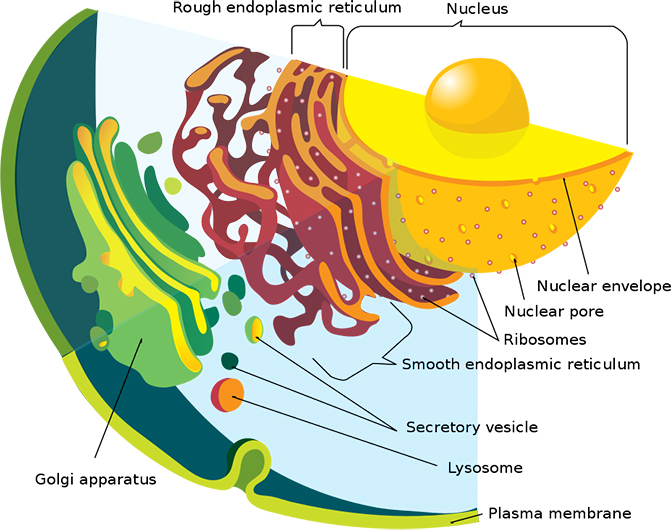 Classic view of a Eukaryote cross section. (Mariana Ruiz/LadyofHats/Wikimedia)
Classic view of a Eukaryote cross section. (Mariana Ruiz/LadyofHats/Wikimedia)
Fusing image data from a library called the Human Protein Atlas and existing maps of protein interactions, the machine learning algorithm was tasked with computing the distances between protein pairs.
The goal was to identify communities of proteins, called assemblies, that co-exist in cells at different scales, from the very small (less than 50 nm) to the very 'large' (more than 1 μm).
One shy of 70 protein communities were classified by the algorithm, which was trained using a reference library of proteins with known or estimated diameters, and validated with further experiments.
Around half of the protein components identified are seemingly unknown to science, never documented in the published literature, the researchers suggest.
In the mix was one group of proteins forming an unfamiliar structure, which the researchers worked out is likely responsible for splicing and dicing newly made transcripts of the genetic code that are used to make proteins.
Other proteins mapped included transmembrane transport systems that pump supplies into and out of cells, families of proteins that help organize bulky chromosomes, and protein complexes whose job it is to make, well, more proteins.
A hefty effort, it's not the first time that scientists have tried to map the inner workings of human cells, though.
Other efforts to create reference maps of protein interactions have yielded similarly mind-boggling numbers and attempted to measure protein levels across tissues of the human body.
Researchers have also developed techniques for visualizing and tracking the interaction and movement of proteins in cells.
This pilot study goes a step further by applying machine learning to cellular microscopy images which locate proteins relative to large cellular landmarks such as the nucleus, and data from protein interaction studies that identify a protein's nearest nano-scale neighbors.
"The combination of these technologies is unique and powerful because it's the first time measurements at vastly different scales have been brought together," says bioinformatician Yue Qin, also of UC San Diego.
In doing so, the Multi-Scale Integrated Cell technique or MuSIC "increases the resolution of imaging while giving protein interactions a spatial dimension, paving the way to incorporate diverse types of data in proteome-wide cell maps," Qin, Ideker and colleagues write.
To be clear, this research is very preliminary: the team focused on validating their method and only looked at the available data from 661 proteins in one cell type, a kidney cell line which scientists have been culturing in the lab for going on five decades.
The researchers plan to apply their newfangled technique to other cell types, says Ideker.
But in the meantime, we'll have to humbly accept we're mere interlopers inside our own cells, capable of understanding a small fraction of the total proteome.
"Eventually we might be able to better understand the molecular basis of many diseases by comparing what's different between healthy and diseased cells," says Ideker.
The study was published in Nature.