For 20 years, experts have relied on the human genome reference as a genetic benchmark for the study of biology, disease, evolution, and more. It's considered one of the greatest scientific achievements of the 21st century.
Now we've got something even better.
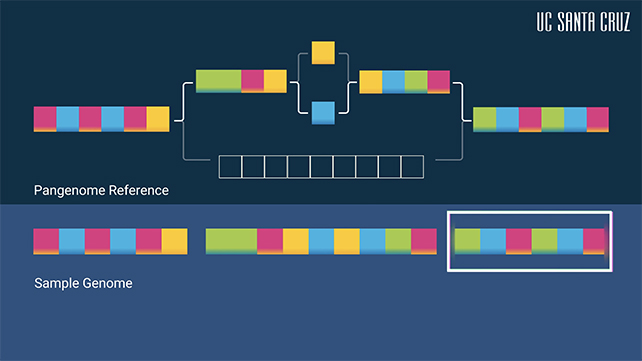
Scientists have just unveiled the first draft of the human pangenome, a much more diverse collection of 47 distinct genomes from different ancestral backgrounds, as opposed to a smaller number of genomes combined into one 'average' reference guide.
It's another major landmark in the field, and will substantially improve attempts to track down the DNA variations that cause all kinds of health conditions, which is handy for tracking down treatments.
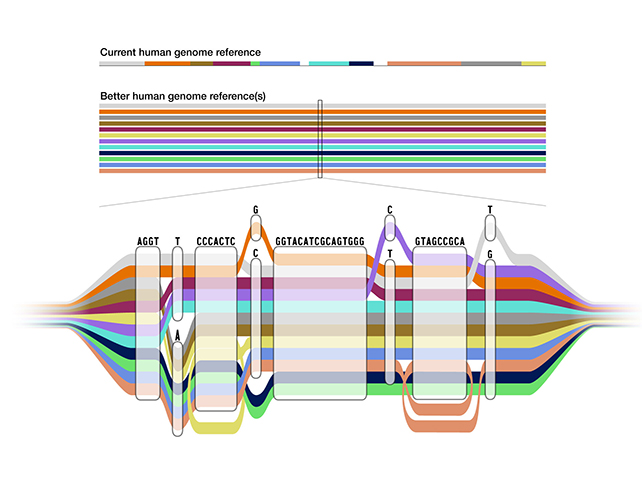
The work has been carried out by a collection of 14 different institutions called the the Human Pangenome Reference Consortium using data taken from the 1,000 Genomes Project catalog. This data was then processed using advanced computational techniques to be more organized and easier to reference.
"We are introducing more diversity and equity into the reference by sampling diverse human beings and including them in this structure that everyone can use," says biomolecular engineer Benedict Paten, from the University of California, Santa Cruz.
"One genome isn't enough to represent everybody – the pangenome will ultimately be something that is inclusive and representative."
Our genomes are more than 99 percent identical. But that single percent of difference accounts for a significant part of makes each individual human being unique (identical twins aside). One single reference point isn't enough to cover all of those variations.
However, the new pangenome can provide several different reference points at once, making it a much more diverse resource for researchers and medical professionals. This helps to remove reference bias, where an analysis is skewed by the data it's relying on.
The new model is 34 percent more accurate at finding small variants, and 104 percent better at finding large (or structural) variants – those that are most difficult to spot – the researchers report.
"Everyone has a unique genome, so using a single reference genome sequence for every person can lead to inequities in genomic analyses," says Adam Phillippy, a genomicist and computer scientist at the National Human Genome Research Institute in Maryland.
"For example, predicting a genetic disease might not work as well for someone whose genome is more different from the reference genome."
Another improvement the pangenome brings with it, thanks to its greater amount and diversity of data, is the ability to distinguish between parental setsof chromosomes – crucial for studying disease inheritance.
This new human pangenome is also going to require new methods of analysis and access, and the team behind it is committed to making it as accessible as possible, so that it can inspire all kinds of future research.
Bear in mind that this is still a first draft. Researchers are hoping to have some 350 genomes in the pangenome by the middle of 2024, making it an exciting time for geneticists, evolutionary biologists, medical researchers, and any other scientist keen to increase our understanding of our own bodies.
"The draft pangenome is an important proof of principle that we hope is going to influence a lot of people and get them thinking about the pangenome and how it might affect their work," says Paten.
"Looking ahead, we see a lot of engagement with other groups – it takes a lot of different people to build something that is going to become a big community resource."
The research has been published in Nature here, here, and here, and in Nature Biotechnology.
