By replacing fluorescent molecules in an existing imaging process with ones that instead scatter light, researchers have revealed a whole new level of dazzling details within our living cells.
The innovative tweak will allow scientists to directly observe molecular behavior over a much longer period, opening a window onto pivotal biological processes like cell division.
"The living cell is a busy place with proteins bustling here and there," explains University of Michigan biomedical engineer Guangjie Cui. "Our superresolution is very attractive for viewing these dynamic activities."
Superresolution is a process for observing incredibly small biological structures. It uses a series of snapshots taken of constellations of fluorescing molecules that highlight select areas of the targeted tissue, eliminating the blurring effect of a flood of diffracted light.
The researchers behind its development won a Nobel prize in 2014. As revolutionary as the process was, the ability of the fluorescing molecules to absorb and then spit back out the required wavelength of light wears out within tens of seconds, which rules out the mapping of longer-duration processes.
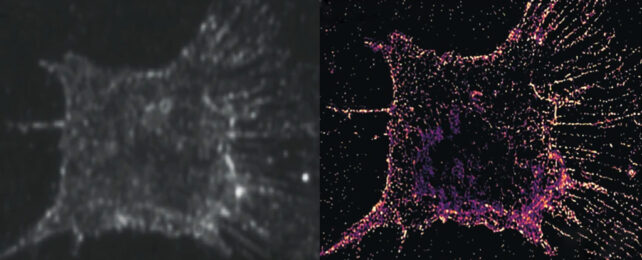
So Cui and colleagues developed a system to detect light scattering off randomly distributed gold nanorods instead, a process that doesn't break down with repeated light exposure. Even though the gold markers are larger than the target structures, imaging multiple differently-angled subsets of the rods and combining the images provides the same highly detailed resolution.
The resulting system allows a staggering 250 hours of continued observations at a resolution of just 100 atoms.
Cui and colleagues then examined the entire process of cellular division with their new PINE nanoscopy, revealing a never-before-seen behavior of actin molecules, down to the individual molecule level.
Actin, the major component of a cell's cytoskeleton, provides cells with structural support and helps facilitate movement within a cell. So these branching filament shaped molecules play a massive role in dividing a cell before pulling it apart into two daughter cells.
Each copy of these cells inherits the same insides, from proteins to DNA, but exactly how this happens has long been a mystery due to the limitations of our visual technology.
Observing 904 actin filaments during the cell division process, Cui and his team could see how individual molecules behaved with each other. They found that when actin molecules are less bound to one another they will expand in search of more links. As each actin reaches its neighbors, it draws other actin molecules close, increasing its network further.
The researchers saw how these small scale movements translated across a larger scale cellular view. Unexpectedly, when actin expands the cell at large actually contracts, whereas it expands when actin contracts. This seems contradictory so the researchers are keen to explore how this opposing motion is occurring.
"We plan to use our method to study how other molecular building blocks organize into tissues and organs," University of Michigan biomedical engineer Somin Lee wrote for The Conversation.
"Our technique could potentially help researchers visualize and, in turn, better understand how molecular defects in tissues and organs may develop into disease."
This research was published in Nature Communications.