As antibiotic-resistant bacteria become increasingly challenging to defeat, consulting those who've been in battle with our ancient foe long before us just makes a whole lot of sense.
Viruses called bacteriophages have been clashing with bacteria since long before we even existed. So researchers from the University of California, San Diego, took a closer look at some of the lesser studied jumbo phages, specifically 201phi2-1, which infects Pseudomonas chlororaphis bacteria.
Named for their large genomes of over 200,000 base pairs long (most phages that infect Pseudomonas have genomes less than 100,000 base pairs), jumbo phages have an arsenal of techniques to counter bacterial defense mechanisms. Previously research found growing a shield around its genetic material was one of these tactics, making these viruses' life cycle unique.
 A cryo-EM representation of the jumbo phage compared to T4 phage virus. (Villa Lab/UCSD)
A cryo-EM representation of the jumbo phage compared to T4 phage virus. (Villa Lab/UCSD)
Separation of genomic material from the rest of the cell contents by encasing it "was previously thought to have evolved only once in the history of life", molecular biologist Thomas Laughlin and colleagues write in their new paper. But it turns out these tiny giants beat us to it.
"It's a different kind of compartment – unlike anything we have ever seen in nature," says biophysicist Elizabeth Villa.
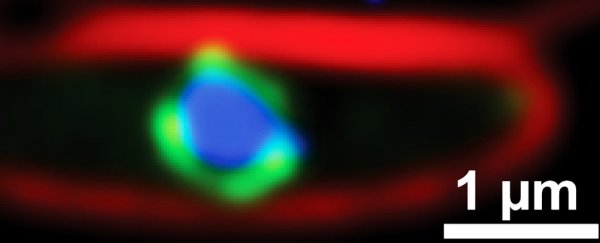
Usually, phages inject their genetic material into the bacteria where it floats freely in the cell's inner goo (cytoplasm) as the virus hijacks the bacteria's equipment to replicate itself. But these jumbo phages construct a separating compartment around their DNA soon after it enters its host, a bit like how our cells have a nucleus to protect our DNA.
This physically prevents the bacteria's CRISPR 'immune system' (yes, it's the same systems we humans have co-opted for our own gene manipulation purposes) and other defensive enzymes from messing with the viral DNA.
Laughlin and their team used cryo-electron microscopy and tomography to examine this compartment down to the atomic scale. The protective casing was constructed from just a single type of protein, which the researchers called chimallin, after an ancient Aztec shield.
 (Villa Lab/UCSD)
(Villa Lab/UCSD)
Above: Micrograph of jumbo phage-infected bacterial cell (left) and nucleus-like compartment outlined in blue (right).
With the help of computer modeling, the researchers found the phage nucleus selectively allows molecules to pass through tiny pores – again similar to how cells control the environment around our genetic material, making this a remarkable example of convergent evolution – when completely unrelated organisms end up with a similar solution to the same biological problem.
"The nuclear pore in eukaryotes is a gigantic, complex structure with very distinctive ways of keeping most proteins out but specifically importing others. What we're probably looking at with the jumbo phage is a dramatically simpler method of solving the same problem," explains biochemist Kevin Corbett. "It's an amazingly creative solution – similar but simpler – to protecting its genome from the outside world by building a wall to separate it from bacterial defenses."
Incredibly, this shield can also grow as the phage's genome replicates. The researchers aren't quite sure how phages manage this yet but suspect the compartment is probably cracking open to allow more chimallin units to join it, which are produced in abundance on infection.
"Now that we know certain phages have a shield, we could give it to other phages and make 'super phages' that are better at phage therapy and overcoming bacterial defenses," says cell biologist Joe Pogliano.
"The first step in that process is understanding the structure of the chimallin protein which makes up the shield, which is one reason this work is so important."
Phage therapy is already being used to successfully treat patients with superbug infections. It is also being considered for customizing our microbiomes when they go out of whack.
With these pesky strains of bacteria refusing to die they are expected to kill 10 million people annually by 2050. So any hints we can get from the enemy of our enemy to better defend ourselves, can't come soon enough.
This research was published in Nature.